Developments from the Shooshtari Lab
Here we display some of the computational tools and machine learning methods that our lab has developed.

scATAC-Explorer: A Single-Cell ATAC-seq Database and Search Tool
scATAC-Explorer is a curated collection of publicly available scATAC-seq (Single Cell Assay for Transposase-Accessible Chromatin using sequencing) datasets. It aims to provide a single point of entry for users looking to study chromatin accessibility at the single-cell level. No standard format for scATAC-seq data exists, so by accessing datasets using scATAC.Explorer, a consistent format is available to users to easily retrieve and integrate multiple datasets together with.
Users can quickly search publicly available datasets using the metadata table, and then download the datasets they are interested in for analysis. Optionally, users can save the datasets for use in applications other than R.
This package will improve the ease of studying and integrating scATAC-seq datasets. Developers may use this package to obtain data for analysis of multiple tissues, diseases, cell types, or developmental stages. It can also be used to obtain data for validation of new algorithms.
https://bioconductor.org/packages/release/data/experiment/html/scATAC.Explorer.html

TMExplorer: A Tumour Microenvironment Single-cell RNAseq Database and Search Tool
TMExplorer (Tumour Microenvironment Explorer) is a curated collection of scRNAseq datasets sequenced from tumours. It aims to provide a single point of entry for users looking to study the tumour microenvironment gene expressions at the single-cell level. Users can quickly search available datasets using the metadata table, and then download the datasets they are interested in for analysis. Optionally, users can save the datasets for use in applications other than R. This package will improve the ease of studying the tumour microenvironment with single-cell sequencing. Developers may use this package to obtain data for validation of new algorithms and researchers interested in the tumour microenvironment may use it to study specific cancers more closely.
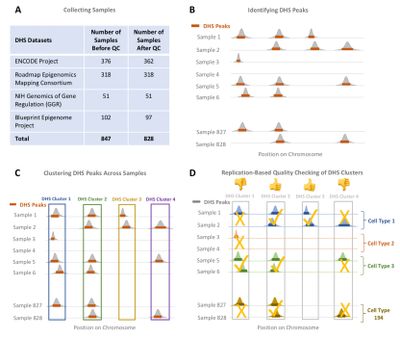
OCHROdb Gene Regulatory Database
OCHROdb is a comprehensive and well-curated database of regions of open chromatin using DNase-I Hypersensitive (DHS) datasets generated by multiple large-scale international projects including REMC, ENCODE, GGR and Blueprint Epigenome Projects. This database contains DHS intensities for 1,455,046 DHS sites aligned over 828 samples. These 828 samples comprise of 194 distinct cell types, including 27 immune cell types, 18 CNS, 9 muscle, 57 cell lines, 10 stem cells, 6 digestive system cell types and several other cell types.

Software for Regulatory Fine Mapping (RegFM)
Regfm is a command line tool that identifies individual regulatory regions in the genome that mediate risk to the disease, and the genes that these regulatory elements control. Regfm uses summary statistics from genome-wide association studies and a list of lead SNPs as the input, and outputs posterior probabilities for each association being mediated by a regulatory effect, for each regulator being the causal risk factor, and for each gene in the locus being pathogenic.

SamSPECTRAL Bioconductor Package
SamSPECTRAL is a Bioconductor package in R capable of clustering large size Flow Cytometry (FCM) data.
http://bioconductor.org/packages/release/bioc/html/SamSPECTRAL.html